Small nuclear ribonucleoprotein polypeptide A
| SNRPA | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | SNRPA, Mud1, U1-A, U1A, Small nuclear ribonucleoprotein polypeptide A | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 182285; MGI: 1855690; HomoloGene: 3380; GeneCards: SNRPA; OMA:SNRPA - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
U1 small nuclear ribonucleoprotein A is a protein that in humans is encoded by the SNRPA gene.[5][6]
Interactions
Small nuclear ribonucleoprotein polypeptide A has been shown to interact with CDC5L.[7]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000077312 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000061479 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Schonk D, van Dijk P, Riegmann P, Trapman J, Holm C, Willcocks TC, Sillekens P, van Venrooij W, Wimmer E, Geurts van Kessel A (January 1991). "Assignment of seven genes to distinct intervals on the midportion of human chromosome 19q surrounding the myotonic dystrophy gene region". Cytogenetics and Cell Genetics. 54 (1–2): 15–9. doi:10.1159/000132946. PMID 1701111.
- ^ "Entrez Gene: SNRPA small nuclear ribonucleoprotein polypeptide A".
- ^ Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI (December 2000). "Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry". The EMBO Journal. 19 (23): 6569–81. doi:10.1093/emboj/19.23.6569. PMC 305846. PMID 11101529.
Further reading
- Hoffman DW, Query CC, Golden BL, White SW, Keene JD (March 1991). "RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins". Proceedings of the National Academy of Sciences of the United States of America. 88 (6): 2495–9. Bibcode:1991PNAS...88.2495H. doi:10.1073/pnas.88.6.2495. PMC 51259. PMID 1826055.
- Nelissen RL, Sillekens PT, Beijer RP, Geurts van Kessel AH, van Venrooij WJ (June 1991). "Structure, chromosomal localization and evolutionary conservation of the gene encoding human U1 snRNP-specific A protein". Gene. 102 (2): 189–96. doi:10.1016/0378-1119(91)90077-O. PMID 1831431.
- Jessen TH, Oubridge C, Teo CH, Pritchard C, Nagai K (November 1991). "Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA". The EMBO Journal. 10 (11): 3447–56. doi:10.1002/j.1460-2075.1991.tb04909.x. PMC 453073. PMID 1833186.
- Nagai K, Oubridge C, Jessen TH, Li J, Evans PR (December 1990). "Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A". Nature. 348 (6301): 515–20. Bibcode:1990Natur.348..515N. doi:10.1038/348515a0. PMID 2147232. S2CID 4336326.
- Sillekens PT, Habets WJ, Beijer RP, van Venrooij WJ (December 1987). "cDNA cloning of the human U1 snRNA-associated A protein: extensive homology between U1 and U2 snRNP-specific proteins". The EMBO Journal. 6 (12): 3841–8. doi:10.1002/j.1460-2075.1987.tb02721.x. PMC 553857. PMID 2962859.
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K (December 1994). "Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin". Nature. 372 (6505): 432–8. Bibcode:1994Natur.372..432O. doi:10.1038/372432a0. PMID 7984237. S2CID 9404488.
- Howe PW, Nagai K, Neuhaus D, Varani G (August 1994). "NMR studies of U1 snRNA recognition by the N-terminal RNP domain of the human U1A protein". The EMBO Journal. 13 (16): 3873–81. doi:10.1002/j.1460-2075.1994.tb06698.x. PMC 395300. PMID 8070414.
- Allain FH, Gubser CC, Howe PW, Nagai K, Neuhaus D, Varani G (April 1996). "Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation". Nature. 380 (6575): 646–50. Bibcode:1996Natur.380..646A. doi:10.1038/380646a0. PMID 8602269. S2CID 4265453.
- Avis JM, Allain FH, Howe PW, Varani G, Nagai K, Neuhaus D (March 1996). "Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding". Journal of Molecular Biology. 257 (2): 398–411. doi:10.1006/jmbi.1996.0171. PMID 8609632.
- Lu J, Hall KB (August 1997). "Tertiary structure of RBD2 and backbone dynamics of RBD1 and RBD2 of the human U1A protein determined by NMR spectroscopy". Biochemistry. 36 (34): 10393–405. doi:10.1021/bi9709811. PMID 9265619.
- Allain FH, Howe PW, Neuhaus D, Varani G (September 1997). "Structural basis of the RNA-binding specificity of human U1A protein". The EMBO Journal. 16 (18): 5764–72. doi:10.1093/emboj/16.18.5764. PMC 1170207. PMID 9312034.
- Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M (September 1998). "Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex". Nature Genetics. 20 (1): 46–50. doi:10.1038/1700. PMID 9731529. S2CID 585778.
- Ferré-D'Amaré AR, Zhou K, Doudna JA (October 1998). "Crystal structure of a hepatitis delta virus ribozyme". Nature. 395 (6702): 567–74. Bibcode:1998Natur.395..567F. doi:10.1038/26912. PMID 9783582. S2CID 4359811.
- Lutz CS, Cooke C, O'Connor JP, Kobayashi R, Alwine JC (December 1998). "The snRNP-free U1A (SF-A) complex(es): identification of the largest subunit as PSF, the polypyrimidine-tract binding protein-associated splicing factor". RNA. 4 (12): 1493–9. doi:10.1017/S1355838298981183. PMC 1369720. PMID 9848648.
- Hetzer M, Mattaj IW (January 2000). "An ATP-dependent, Ran-independent mechanism for nuclear import of the U1A and U2B" spliceosome proteins". The Journal of Cell Biology. 148 (2): 293–303. doi:10.1083/jcb.148.2.293. PMC 2174293. PMID 10648562.
- Klein Gunnewiek JM, Hussein RI, van Aarssen Y, Palacios D, de Jong R, van Venrooij WJ, Gunderson SI (March 2000). "Fourteen residues of the U1 snRNP-specific U1A protein are required for homodimerization, cooperative RNA binding, and inhibition of polyadenylation". Molecular and Cellular Biology. 20 (6): 2209–17. doi:10.1128/MCB.20.6.2209-2217.2000. PMC 110837. PMID 10688667.
- Varani L, Gunderson SI, Mattaj IW, Kay LE, Neuhaus D, Varani G (April 2000). "The NMR structure of the 38 kDa U1A protein - PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein". Nature Structural Biology. 7 (4): 329–35. doi:10.1038/74101. PMID 10742179. S2CID 13432764.
- Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI (December 2000). "Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry". The EMBO Journal. 19 (23): 6569–81. doi:10.1093/emboj/19.23.6569. PMC 305846. PMID 11101529.
- Simmons A, Aluvihare V, McMichael A (June 2001). "Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators". Immunity. 14 (6): 763–77. doi:10.1016/S1074-7613(01)00158-3. PMID 11420046.
- v
- t
- e
-
 1aud: U1A-UTRRNA, NMR, 31 STRUCTURES
1aud: U1A-UTRRNA, NMR, 31 STRUCTURES -
 1cx0: HEPATITIS DELTA VIRUS RIBOZYME
1cx0: HEPATITIS DELTA VIRUS RIBOZYME -
 1drz: U1A SPLICEOSOMAL PROTEIN/HEPATITIS DELTA VIRUS GENOMIC RIBOZYME COMPLEX
1drz: U1A SPLICEOSOMAL PROTEIN/HEPATITIS DELTA VIRUS GENOMIC RIBOZYME COMPLEX -
 1dz5: THE NMR STRUCTURE OF THE 38KDA U1A PROTEIN-PIE RNA COMPLEX REVEALS THE BASIS OF COOPERATIVITY IN REGULATION OF POLYADENYLATION BY HUMAN U1A PROTEIN
1dz5: THE NMR STRUCTURE OF THE 38KDA U1A PROTEIN-PIE RNA COMPLEX REVEALS THE BASIS OF COOPERATIVITY IN REGULATION OF POLYADENYLATION BY HUMAN U1A PROTEIN -
 1fht: RNA-BINDING DOMAIN OF THE U1A SPLICEOSOMAL PROTEIN U1A117, NMR, 43 STRUCTURES
1fht: RNA-BINDING DOMAIN OF THE U1A SPLICEOSOMAL PROTEIN U1A117, NMR, 43 STRUCTURES -
 1m5k: Crystal structure of a hairpin ribozyme in the catalytically-active conformation
1m5k: Crystal structure of a hairpin ribozyme in the catalytically-active conformation -
 1m5o: Transition State Stabilization by a Catalytic RNA
1m5o: Transition State Stabilization by a Catalytic RNA -
 1m5p: Transition State Stabilization by a Catalytic RNA
1m5p: Transition State Stabilization by a Catalytic RNA -
 1m5v: Transition State Stabilization by a Catalytic RNA
1m5v: Transition State Stabilization by a Catalytic RNA -
 1nu4: U1A RNA binding domain at 1.8 angstrom resolution reveals a pre-organized C-terminal helix
1nu4: U1A RNA binding domain at 1.8 angstrom resolution reveals a pre-organized C-terminal helix -
 1oia: U1A RNP DOMAIN 1-95
1oia: U1A RNP DOMAIN 1-95 -
 1sj3: Hepatitis Delta Virus Gemonic Ribozyme Precursor, with Mg2+ Bound
1sj3: Hepatitis Delta Virus Gemonic Ribozyme Precursor, with Mg2+ Bound -
 1sj4: Crystal structure of a C75U mutant Hepatitis Delta Virus ribozyme precursor, in Cu2+ solution
1sj4: Crystal structure of a C75U mutant Hepatitis Delta Virus ribozyme precursor, in Cu2+ solution -
 1sjf: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in Cobalt Hexammine solution
1sjf: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in Cobalt Hexammine solution -
 1u6b: CRYSTAL STRUCTURE OF A SELF-SPLICING GROUP I INTRON WITH BOTH EXONS
1u6b: CRYSTAL STRUCTURE OF A SELF-SPLICING GROUP I INTRON WITH BOTH EXONS -
 1urn: U1A MUTANT/RNA COMPLEX + GLYCEROL
1urn: U1A MUTANT/RNA COMPLEX + GLYCEROL -
 1vbx: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in EDTA solution
1vbx: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in EDTA solution -
 1vby: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaiton, and Mn2+ bound
1vby: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaiton, and Mn2+ bound -
 1vbz: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in Ba2+ solution
1vbz: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in Ba2+ solution -
 1vc0: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in Imidazole and Sr2+ solution
1vc0: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in Imidazole and Sr2+ solution -
 1vc5: Crystal Structure of the Wild Type Hepatitis Delta Virus Gemonic Ribozyme Precursor, in EDTA solution
1vc5: Crystal Structure of the Wild Type Hepatitis Delta Virus Gemonic Ribozyme Precursor, in EDTA solution -
 1vc6: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Product with C75U Mutation, cleaved in Imidazole and Mg2+ solutions
1vc6: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Product with C75U Mutation, cleaved in Imidazole and Mg2+ solutions -
 1vc7: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in Sr2+ solution
1vc7: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutation, in Sr2+ solution -
 1zzn: Crystal structure of a group I intron/two exon complex that includes all catalytic metal ion ligands.
1zzn: Crystal structure of a group I intron/two exon complex that includes all catalytic metal ion ligands. -
 2nz4: Structural investigation of the GlmS ribozyme bound to its catalytic cofactor
2nz4: Structural investigation of the GlmS ribozyme bound to its catalytic cofactor -
 2oih: Hepatitis Delta Virus gemonic ribozyme precursor with C75U mutation and bound to monovalent cation Tl+
2oih: Hepatitis Delta Virus gemonic ribozyme precursor with C75U mutation and bound to monovalent cation Tl+ -
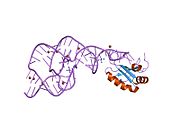 2oj3: Hepatitis Delta Virus ribozyme precursor structure, with C75U mutation, bound to Tl+ and cobalt hexammine (Co(NH3)63+)
2oj3: Hepatitis Delta Virus ribozyme precursor structure, with C75U mutation, bound to Tl+ and cobalt hexammine (Co(NH3)63+) -
 2u1a: RNA BINDING DOMAIN 2 OF HUMAN U1A PROTEIN, NMR, 20 STRUCTURES
2u1a: RNA BINDING DOMAIN 2 OF HUMAN U1A PROTEIN, NMR, 20 STRUCTURES
 | This article on a gene on human chromosome 19 is a stub. You can help Wikipedia by expanding it. |
- v
- t
- e















































